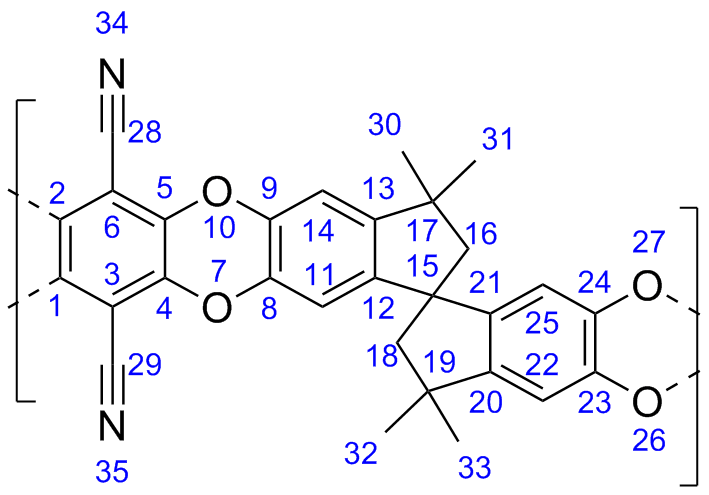
This database is a collection of the molecular models used in some publications of the Colina Research Group at the University of Florida.
Select a molecule by navigating the drop down menu below to obtain information on the force field functional forms and parameters used in published articles. For each model, you may download data files containing all necessary molecular modeling information. If using any of the information provided in this database, please cite the corresponding paper, as listed on the webpage for each model.
Please select an option from the drop down menu to access the force field information:
Contributors
- Coray Colina - University of Florida
- Kyle E. Hart - Penn State
- Lauren J. Abbott - Penn State
- Michael E. Fortunato - University of Florida
- Akshay Mathavan - University of Florida
- Akash Mathavan - University of Florida
- Farhad Ramezanghorbani - University of Florida
- Ping Lin - University of Florida
- Alexander Demidov - University of Florida
Funding
Funding for this project is provided by the U.S. Department of Energy, Office of Science, Basic Energy Sciences, under Award DE-FG02-17ER16362, as part of the Computational Chemical Sciences Program. Funding for this project was initially provided by the National Science Foundation Grant (DMR0908781 and DMR1310258).
Under the '(Polymers)' is the zipped folder with LAMMPS data files of polymer models built from the corresponding monomers. Provided in this download is the force field information used for this coarse-grained model, including: GROMACS input files (.mdp, .gro, .itp) and a Martini parameter file (.itp).
Citation
Please site the following article when using this information:
- Hart, K. E.; Colina, C. M. “Estimating gas permeability and permselectivity of microporous polymers.” J. Membr. Sci 2014, 468, 259-268
Parameters
A naming scheme is used in this work for each united atom bead. The naming scheme used is:
[element][type][hydrogens]
[element]: the chemical element of the atom
[type]: the chemical environment of the atom,
A = Aromatic, H = Tetrahedral, L = Linear, P = Trigonal Planar, S = Shared Aromatic, K = Ketone
[hydrogens]: the number of bonded hydrogen atoms in the pseudo atom
For example, CH2 is a tetrahedral carbon unit atom bead with two associated hydrogens. An “L” preceding a Linking atom type designates an atom being used in a Polymatic bonding step.
LAMMPS Parameters
Detailed information for the format of LAMMPS data files may be found in the documentation. The specific functional forms used in these data files are the following:
References
Download information
The detailed description of the zipped content can be found in this archive:
Citation
Please site the following article when using this information:
Please report your issue below and we will work on fixing it as soon as possible.
Issue:
Your email was successfully sent.
Error: There was an issue sending your email, please try again.